My research has focused on the application of computational biology and machine learning tools and approaches to solve biological problems, for drug discovery and synthetic biology. The application of computational thinking to these problems has the potential to revolutionize the way we approach them, and to provide new insights and solutions that are unexpected or innovative. Continuous innovation into computational methods to solve challenges in biology – be it drug discovery, synthetic biology, or protein design – is essential to the future of these fields.
Drug repositioning and a new way of looking at expression profiles
One of the most challenging problems in drug discovery is the identification of new uses for existing drugs. This is known as drug repositioning, and its challenges arise from a need to the biological mechanisms underlying the disease of interest and the drug’s mechanism of action.
The existence of extensive libraries of drug perturbations against cell lines and biological systems, measures through omics readout such as transcriptomics, allows one to develop computational tools that harness these data and predict novel uses for existing molecules.
I have developed the DRUID R package (see here) that can address this challenge. The major feature of DRUID is how we look at the expression profile of a system in response to a drug perturbation: by taking in cues from Natural Language Processing, we look at an expression profile as a “document”, where the expression of each gene is considered to be a “word” in the document. This abstraction allows us to use a variety of NLP tools to analyze the expression profile, such as the term frequency and inverse document frequency statistics for words in a corpus:
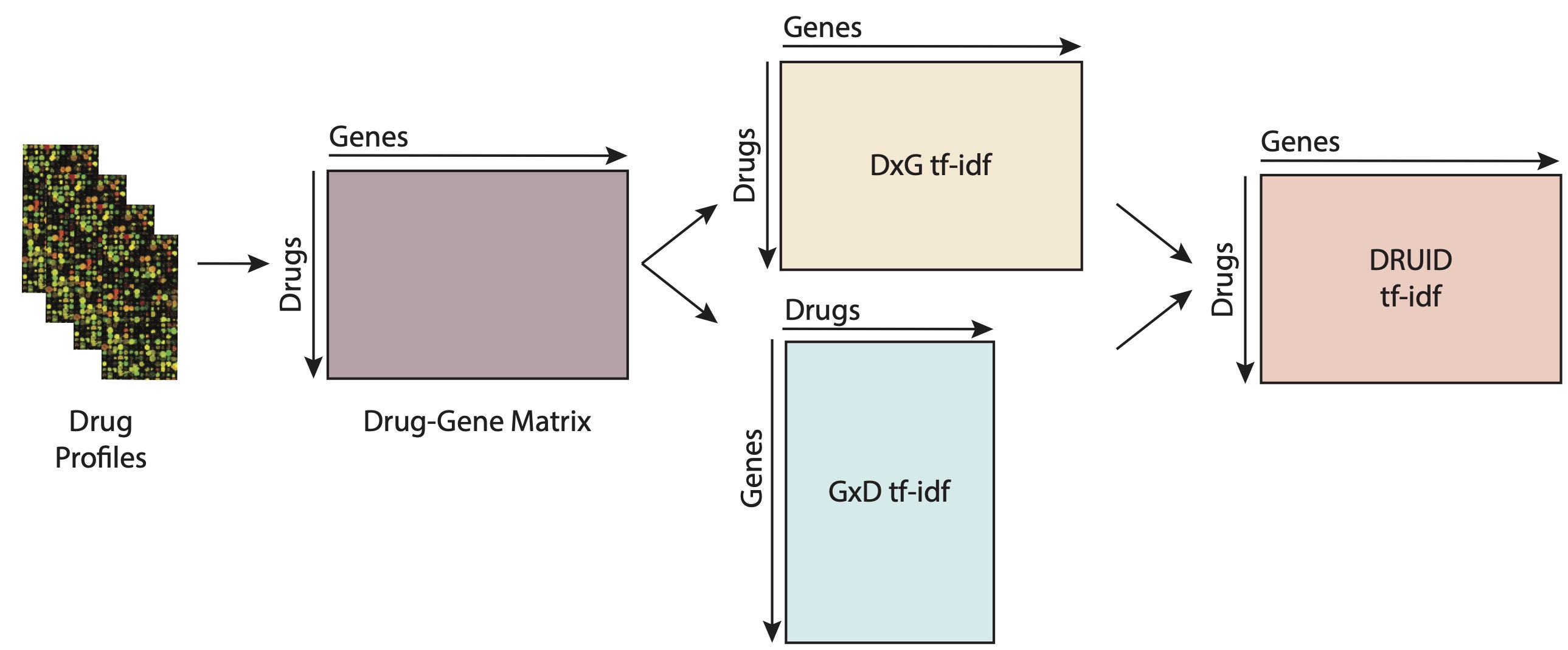
This framework showed some success in in vitro validation studies, where we looked for molecules that could impact the growth of lung cancer cells. We found a few molecules that were not known to have an effect on lung cancer, but that showed a strong effect in our in vitro assays.
Differential network analysis for the identification of key topological differences between biological states
When we measure the impact of a perturbation on a biological system, we tend to use at the individual biomolecules in isolation, be it for target discovery or the deconvolution of the mechanism of action of a drug. However, biological systems are complex networks of interactions between biomolecules, and the impact of a perturbation on a system can be better understood by looking at the network as a whole.
Statistical and mathematical approaches like network inference and system identification allows us to identify the network topology of a biological system. Then, as you look across different states, you can infer the key topological differences between these states. This is the basis of differential network analysis, and I have developed the DiffNet R package (see here) to perform this type of analysis.
Integrating these approaches with tools such as DRUID, you can build a comprehensive computational platform that will give you novel insights and a new way of looking at biological systems.
Synthetic biology and the design of toehold switches
Synthetic biology is a field that aims to design and build new biological systems and devices. One of the most exciting applications of synthetic biology is the design of genetic circuits that can sense and respond to environmental cues. One of the most promising genetic circuits is the toehold switch, a small RNA molecule that can be designed to bind to a target mRNA and regulate its translation.
Work that we did at the Wyss, using a combination of Natural Language Processing and Convolutional Neural Networks, allowed us to design a novel computational platform, STORM, that can predict the activity of a toehold switch based only on its sequence.
